Determining the Current Phytophthora Sojae Population and Status of Variety Resistance
Organization awarded: University of Minnesota
Principal Investigator: Megan McCaghey
Cooperators: Kathleen K. Markham, Linnea Johnston, Jane Fenske-Newbart, Crystal Floyd, Cathy Johnson, Senyu Chen, Carol Groves, Damon Smith, Dean Malvick and Megan McCaghey

Summary Content
Over the past five growing seasons, parts of Minnesota have experienced above-average soil moisture (Fig. 1), potentially promoting the development of Phytophthora sojae, a soil-borne pathogen that attacks soybeans at all growth stages. Planting resistant soybean varieties is a key management strategy. However, resistance genes (Rps) must align with the P. sojae pathotypes in the field to be effective. In this study, Dr. Kathleen Markham aims to survey P. sojae pathotypes in Minnesota and validate a molecular diagnostic method. Results will help recommend soybean varieties with potentially effective Rps genes that are more aligned to local pathotypes, enhancing disease resistance.
We are conducting a pathotype survey using soil and plant tissue collected during the 2023–2025 growing seasons. We have received 97 samples from 21 counties (Fig. 2) and cultured 30 isolates using a McCaghey Lab-optimized baiting method. Twenty-five isolates tested positive via PCR for Phytophthora-specific, P. sojae-specific, and P. sojae avirulence (Avr) genes, confirming their identity.
Findings
As pathotyping continues, we will provide results and variety recommendations to growers who submitted samples. Based on preliminary molecular results (Table 1), Rps3a or Rps6 varieties are predicted to be resistant to many of the new isolates, while Rps1a, Rps1c, and Rps1k are predicted to offer little resistance. Notably, Rps1a, Rps1c, and Rps1k—once common—are becoming less effective, while Rps3a, Rps6, and Rps11 may be more effective (McCoy et al., 2023).
We analyzed 2019–2023 UMN Soybean Variety Trial data to evaluate whether deployed Rps genes align with current field pathotypes. Stacked gene combinations (e.g., Rps1a + Rps3a) have increased (Fig. 3), suggesting a strategy to address complex pathotypes. However, use of Rps6 is declining, while Rps3a is increasing. Potentially ineffective genes (Rps1a, Rps1c, Rps1k) still appear frequently in trials, suggesting room for improvement in variety selection.
Our survey, combined with variety trial analysis, will guide growers toward soybean varieties that can potentially resist local P. sojae pathotypes. Outreach will include extension materials, meetings, and reports. The project aims to reduce yield loss by delivering rapid diagnostics and effective variety recommendations, while also exploring partial resistance to improve long-term management strategies.
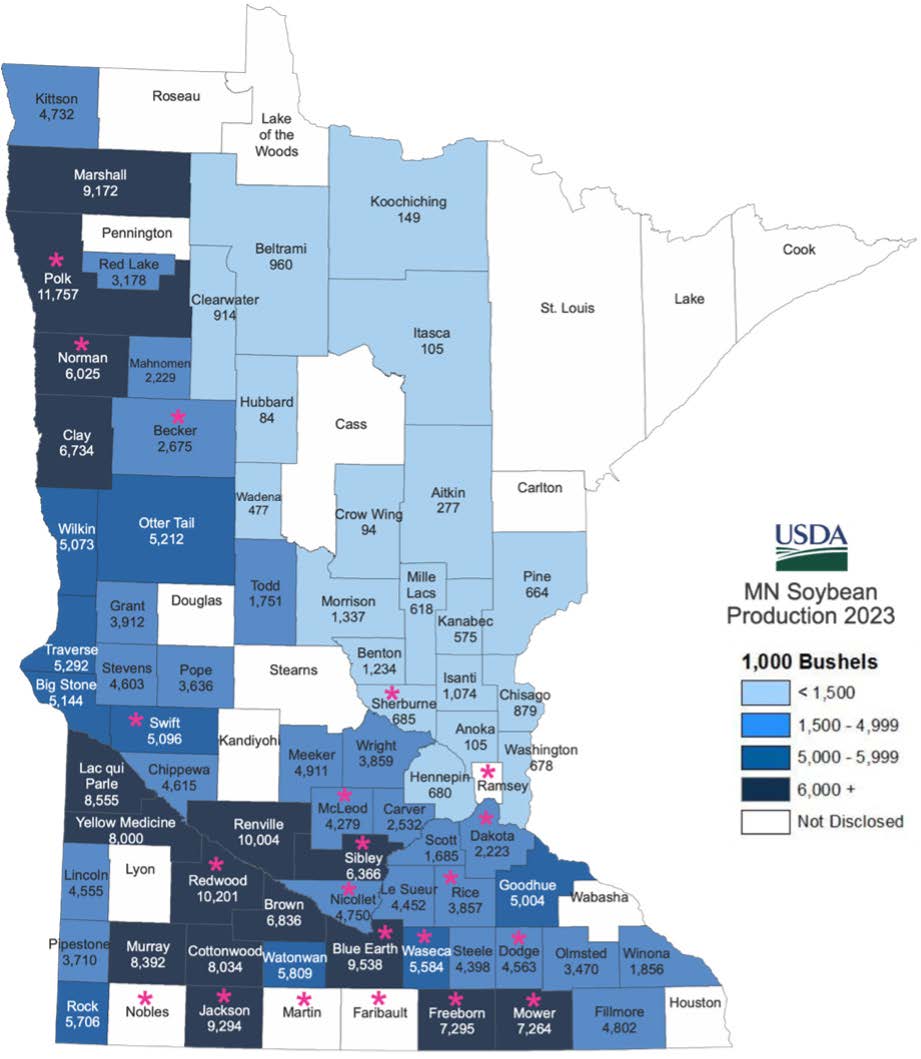
Figure 1. Soil and tissue samples collected from various MN fields in 2023-2025. Pink asterisk represents counties from which we have soil and tissue samples. Map of MN soybean production in 2023, showing number of bushels produced per county (Source: USDA)
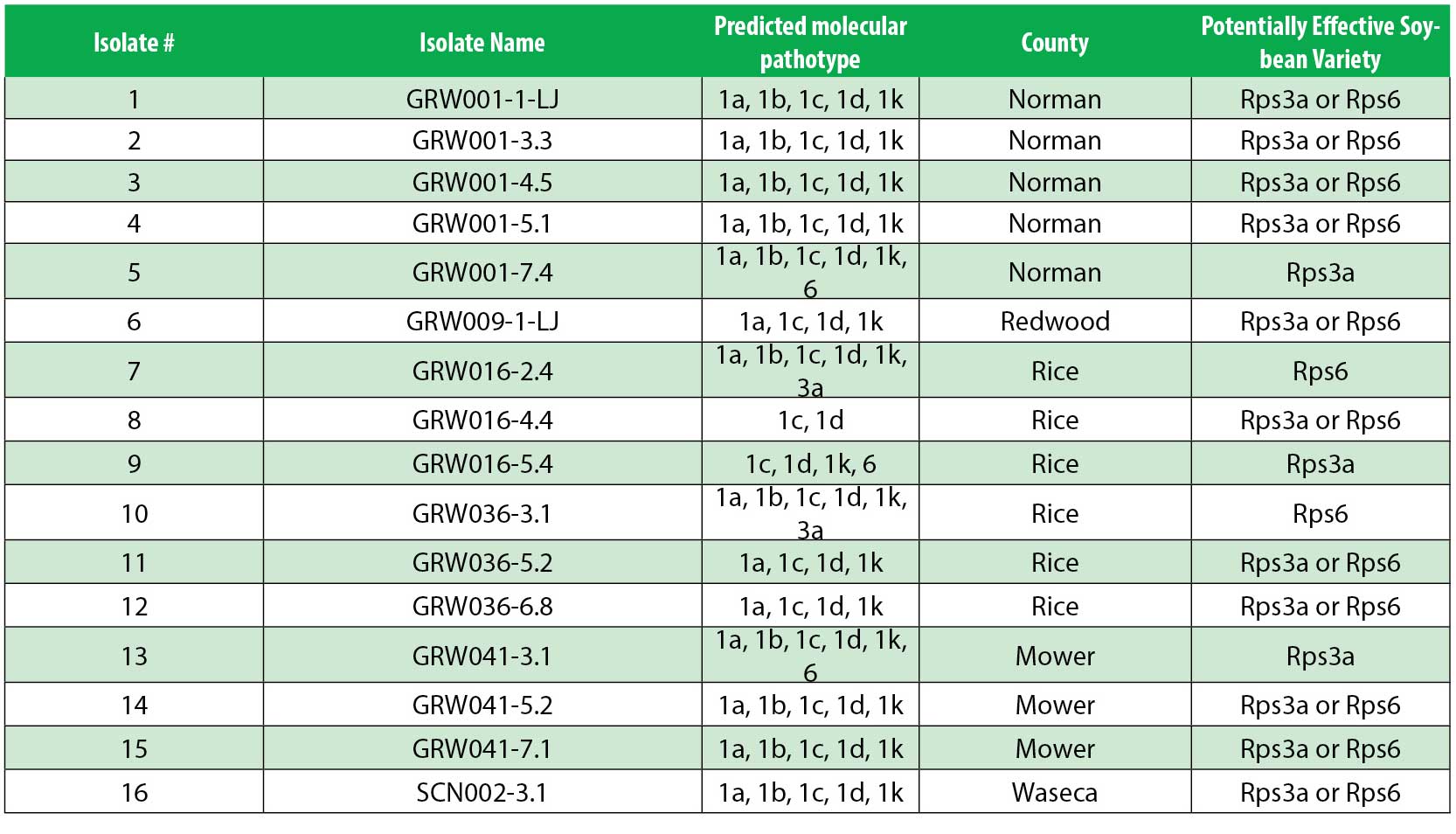
Table 1. Preliminary identification of 16 (of 25) P. sojae-confirmed isolates recovered from soil collected around MN in 2023-2025. There are 6 unique pathotypes across 5 counties thus far.
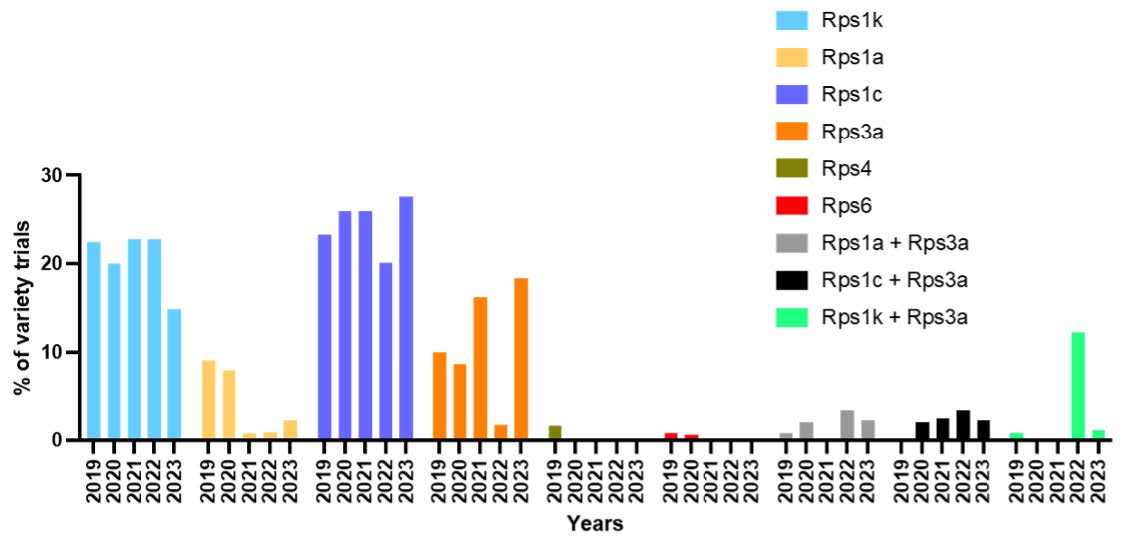
Figure 2. Frequency of Rps genes in the UMN Soybean Crop Variety Trials in years 2019-2023.

